Coral Reef Classification: 2016 REU

Coral reefs are large, diverse, and extremely important ecosystems spread throughout the world’s oceans. Unfortunately, in the past year a significant portion of this ecosystem is suffering in recent years, heavily due to rising ocean temperatures. Over 90% of the northern Great Barrier reef is bleached and over 20% has died. Bleaching is when the coral releases its symbiotic algae used for food. Ecologists are interested in using high resolution spatial and temporal data to further determine behavior and relationships of different corals, e.g. symbiotic relationships, competition between species, and growth rates. This is often determined through selective statistical sampling of small areas, or large area image collection using human divers or AUVs at multiple time steps. This second method of large scale image collection poses the interesting challenge of dealing with large quantities of unlabeled natural data. As images are not inherently useful for ecologists to determine the above mentioned relationships, the data must be semantically classified into the different taxonomic representations of the coral. Hand labeling of imagery is extremely time consuming, at nearly a 100:1 ratio between labeling and data collection time, which unfortunately makes large scale hand segmentation and classification unfeasible. The focus of this REU was to automate this procedure.
Our project in the 2016 E4E REU program focused on automating semantic segmentation and classification of orthomosaic and pointcloud coral imagery in collaboration with the Sandin Lab for spatial coral ecology at Scripps Institute of Oceanography. Sandin Lab has dove throughout the world and collected dozens of datasets (and plans on expanding through the 100 Island Challenge in which the image 100 different island in a single field season), unfortunately under 0.1% of this data is labeled.This effectively became an applied machine learning problem. Our summer project applied several machine learning algorithms to label the imagery and point clouds automatically, specifically SVMs, FF-ANN, and random forests trained on manual features, and fully convolutional neural networks were applied to the point cloud and image datasets. Further work is being pursued to increase accuracy.
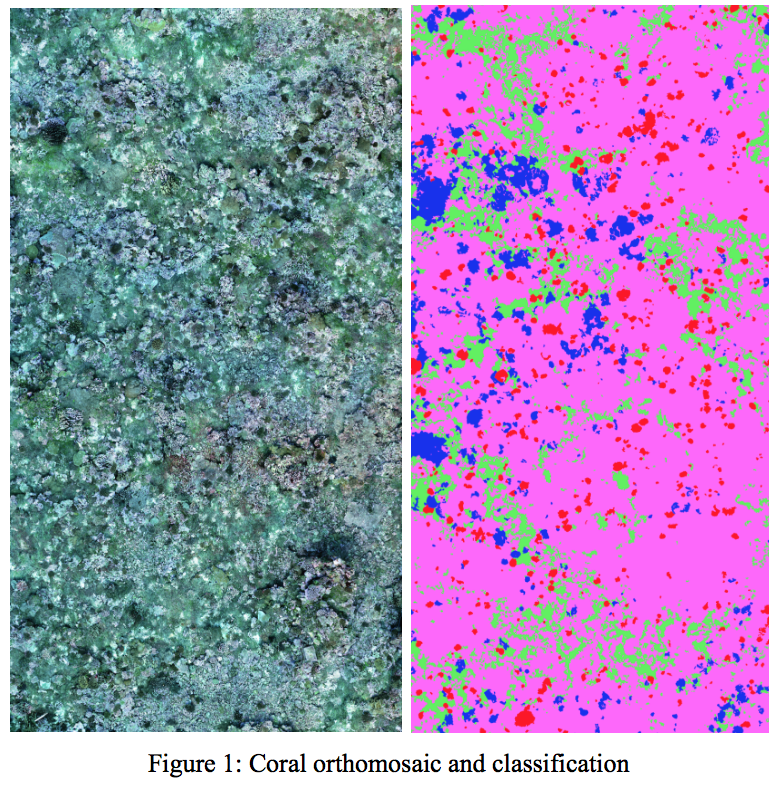
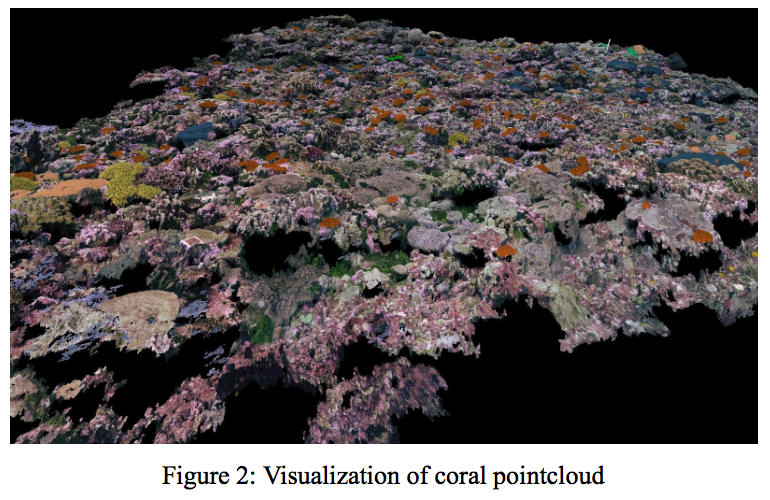
Image and point-cloud accuracy were measured at a per pixel (point) level for evaluation. Percent accuracy is (#correct pixels)/(total #pixel) for evaluation. This does not take into account various class weightings, all pixels are weighted the same.
4 class coral:
| Method | Test Accuracy |
|---|---|
| Unet with augmentation | 72.0% |
| FCN with augmentation and early stopping | 78.2% |
| Unet with augmentation and class weighting | 82.4% |
| Segnet w/out augmentation | 82.6% |
| Segnet with augmentation | 88.9% |
22 class coral:
| Method | Test Accuracy |
|---|---|
| Segnet with augmentation | 76.2% |
| Random Forest with local features | 60.6% |
| Pointclouds using ANN | 67.8% |
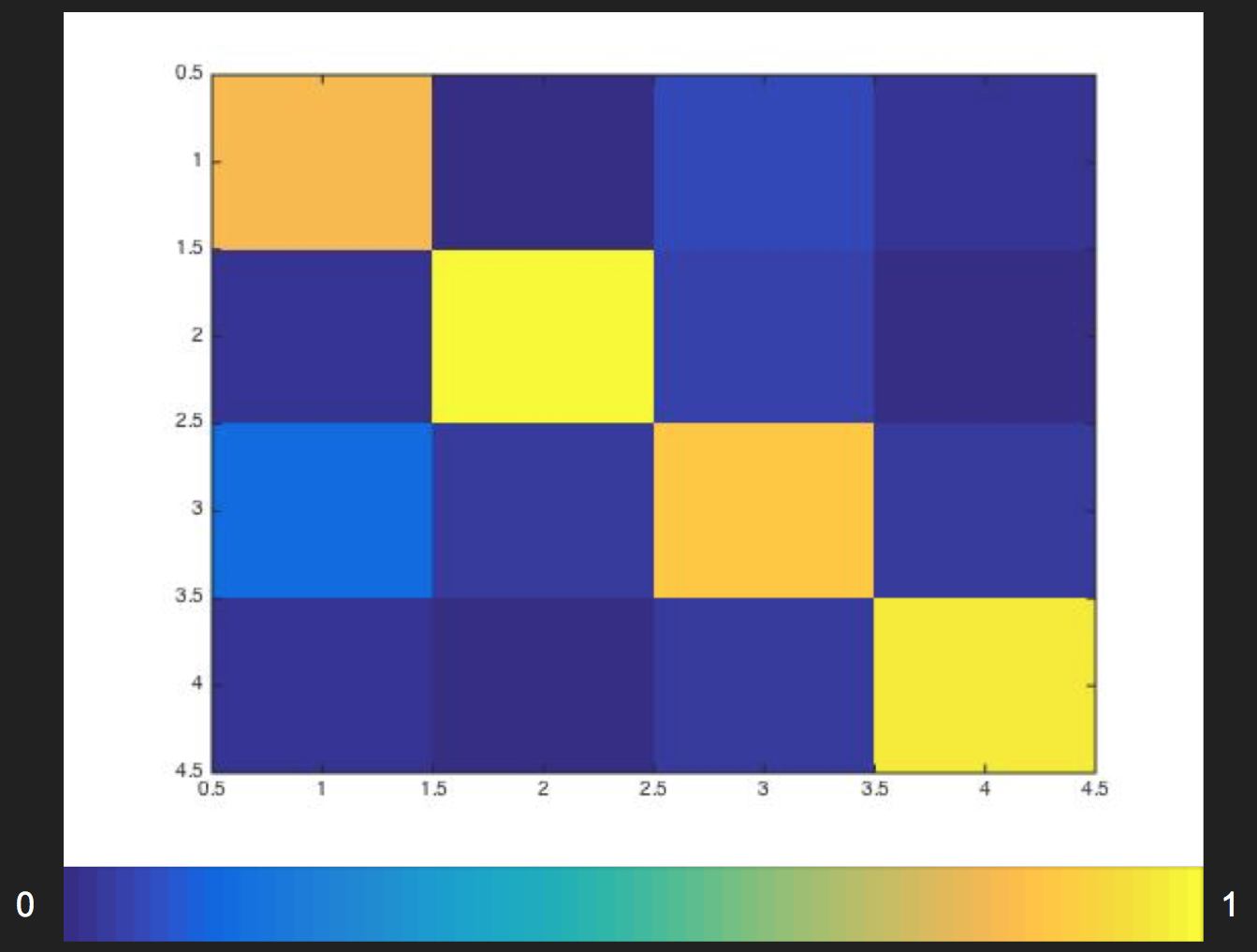
Figure 3: Segnet with augmentation confusion matrix
Segnet with augmentation confusion matrix, ratio:
| Confusion Matrix | Predicted Class 1 | Predicted Class 2 | Predicted Class 3 | Predicted Class 4 |
|---|---|---|---|---|
| True Class 1 | 0.732 | 0.002 | 0.024 | 0.017 |
| True Class 2 | 0.015 | 0.976 | 0.039 | 0.013 |
| True Class 3 | 0.235 | 0.019 | 0.902 | 0.234 |
| True Class 4 | 0.016 | 0.003 | 0.035 | 0.950 |
This has been a very interesting project, unfortunately it has not yet fully automated coral classification at full taxonomical resolution. Accuracy was sufficient on the four class system that collaborators at Scripps were satisfied, however that is not matching their classification scheme. I will ask them if they may be able to use this to pre-classify, if that would be useful. I am hopeful with further experimentation automated image classification is feasible.
Future approaches are applying deeper and selectively trained models with varying levels of supervision (grouping several classes into larger morphologies, before getting to fine taxonomical representation or an initial segmentation into background and foreground classes, before finer segmentation). The low class problem worked extremely well, however jumping straight to full taxonomical resolution performed very poorly. In between steps may fix this. Point Cloud classification experiments performed surprisingly well given the lack of structural labeling, using methods structural methods like fixed-point or MRF and CRF would produce far better results.
